Exploring terra incognita of cognitive science: Lateralization of gene expression at the frontal pole of the human brain
Abstract
Background. Rostral prefrontal cortex, or frontopolar cortex (FPC), also known as Brodmann area 10 (BA10), is the most anterior part of the human brain. It is one of the largest cytoarchitectonic areas of the human brain that has significantly increased its volume during evolution. Anatomically the le (BA10L) and right (BA10R) parts of FPC show slight asymmetries and they may have distinctive cognitive functions. Objective. In the present study, we investigated differential expression of the transcriptome in the le and right parts of BA10.
Design. Postmortem samples of human brain tissue from fourteen donors (male/ female without history of psychiatric and neurological diseases, mean age 39.79±3.23 years old, mean postmortem interval 12.10±1.76 h) were obtained using the resources of three institutions: the Partner Institute of Computational Biology of Chinese Academy of Sciences, the Max Planck Institute for Evolutionary Anthropology, and NIH Neuro-BioBank.
Results. By using a standard RNA-sequencing followed by bioinformatic analysis, we identified 61 genes with differential expression in the le and right FPC. In general, gene expression was increased in BA10R relative to BA10L: 40 vs. 21 genes, respectively. According to gene ontology analysis, the majority of up-regulated genes in BA10R be- longed to the protein-coding category, whereas protein-coding and non-coding genes were equally up-expressed in BA10L. Most of the up-regulated genes in BA10R were involved in brain plasticity and activity-dependent mechanisms also known for their role in the hippocampus. 24 out of 30 mental disorder-related genes in the dataset were disrupted in schizophrenia. No such a wide association with other mental disorders was found.
Conclusion. Discovered differences point at possible causes of hemispheric asymmetries in the human frontal lobes and at the molecular base of higher-order cognitive processes in health and disease.
Received: 25.08.2017
Accepted: 11.09.2017
Themes: Cognitive psychology
PDF: http://psychologyinrussia.com/volumes/pdf/2017_3/psych_3_2017_17.pdf
Pages: 231-247
DOI: 10.11621/pir.2017.0316
Keywords: neuropsychology, frontopolar cortex, human cerebral asymmetry, yakovlevian torque, RNA transcriptome, sequencing, schizophrenia, attention
Introduction
Rostral prefrontal cortex, or frontopolar cortex (FPC), also known as Brodmann area 10 (BA10), is the most anterior part of the human brain. This area extended its complexity during hominid evolution by e.g. considerable increase in neuron numbers, a specific increase in connectivity and a dramatic increase in size: gibbon – 0.2 cm3(0.2%), bonobo – 2.8 cm3(0.7%), human – 14.2 cm3(1.2% of the whole brain volume)(Semendeferi, Armstrong, Schleicher, Zilles, & Van Hoesen, 2001). Functional neuroimaging studies of prefrontal cortex showed strong hemodynamic changes in this area under different conditions, from the simplest to the most complex tasks, such as language understanding or contemplating future actions, in accordance with internal goals and plans(Burgess, Dumontheil, & Gilbert, 2007; Miller, & Cohen 2001). Together with clinical observations, it appears that rostral prefrontal cortex, considered as a whole, supports higher cognitive processes such as planning, decision-making, retrieval of memories, establishing logical links and multi-tasking that allows performance of mental and physical activities at the same time or in close temporal succession (Braver, & Bongiolatti, 2002; Penfield, & Evans 1935; Ramnani, & Owen, 2004). In general, the left and right hemispheres of the human brain are anatomically asymmetric (LeMay, 1999)andprobably support different cognitive functions. A number of studies suggests a particular role of the right FPC in self-referential rather than other-referential encoding (Craik et al., 1999; Christoff, & Gabrieli, 2000; Soch et al., 2016)and in understanding of concealed, or indirect meaning as in metaphoric speech, humor, irony and sarcasm (see however Forgacs, Lukács, & Pléh, 2014). The functional differences are echoed by anatomical data. Although basic mechanisms of language perception and production, traditionally considered as the differentia specificaof Homo sapiens sapiens, are localized in the left hemisphere, the right frontopolar area (BA10R) has a larger volume and demonstrates more rapid growth, both in anthropogenesis and in early ontogenesis (Hrvoj-Mihic, Bienvenu, Stefanacci, Muotri, & Semendeferi, 2013). This can be related to the Yakovlevian Torque phenomenon, in which frontal structures anterior to the right Sylvian fissure are ‘torqued forward’ relative to their counterparts on the left. The left occipital lobe is also splayed across the midline and skews the interhemispheric fissure in a rightward direction. First described by P.I Yakovlev, this phenomenon has been supported by fragmented paleoneurological findings (Toga, & Thompson, 2003).
Although hemispheric lateralization is not a feature unique to humans (Halpern, Gunturkun, Hopkins, & Rogers, 2005), the most publications on this asymmetry is dedicated to the anatomy, physiology and clinical pathology of the human brain (Herbert et al., 2005; Pujol et al., 2002; Toga, & Thompson, 2003). For example, cognitive disorders and psychiatric diseases such as schizophrenia, autism and dyslexia are accompanied by disturbances in brain asymmetry (Carper, Treiber, DeJesus, & Muller, 2016;Renteria, 2012; Y. Sun, Chen, Collinson, Bezerianos, & Sim, 2017).
Previous research has shown that the human genome produces a consistent molecular architecture in the cortex, despite millions of genetic differences across individuals and races (Colantuoni et al., 2011). However, molecular mechanisms leading to development of brain asymmetry in the adult human brain remain, with a few exceptions, underexplored (Kang et al., 2011; Karlebach, & Francks, 2015). In particular, molecular bases of the functional differences between distinct brain regions are either unclear (Pletikos et al., 2014)and disputed (T. Sun, Collura, Ruvolo, & Walsh, 2006), or complicated for subsequent analysis (Hawrylycz et al., 2012). This is especially true with respect to the evolutionary new prefrontal cortex.
In the present study, we applied the common RNA-Seq technique together with Gene Ontology (GO) analysis to investigate differential expression of the transcriptome in the left and right parts of Brodmann area 10 (BA10L and BA10R), i.e. around the poles of the human brain which show a significant diversity in anatomical and neuropsychological features. Understanding the differences in the transcriptome patterns of these most rostral cortical regions may have important basic and clinical relevance.
Method
Material and methods
Human brain tissue
Postmortem samples of human brain tissue from FPC areas in the left and right hemisphere (BA10L and BA10R) from fourteen donors were obtained using the resources of three institutions: the Partner Institute of Computational Biology of Chinese Academy of Sciences, the Max Planck Institute for Evolutionary Anthropology,and NIH NeuroBioBank. This sampling consisted of seven BA10L samples and seven BA10R samples from male/female individuals without known history of psychiatric and neurological diseases. Mean age 39.79±3.23 years old, mean postmortem interval 12.10±1.76 h (mean±SD).
RNA extraction
Isolation of total RNA from brain tissue samples was carried out using the Trizol reagent (Thermo Fisher Scientific, USA) by the standard technique. The quantity of total RNA was determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific, USA). RNA integrity number (RIN) was assessed by BioAnalyzer 2100 (Agilent Technologies, USA) and RNA 6000 Nano kit (Agilent Technologies, USA). The RIN ranged from 6.1 to 9.3 for all samples.
Library preparation and sequencing
14 cDNA libraries for sequencing were constructed using 10 μg of RNA per sample and the mRNA-Seq Sample Prep Kit (Illumina, USA) according the manufacturer’s protocol. The final library met all quality metrics as defined by Illumina, and library quantization was performed on an Agilent 2100 Bioanalyzer with a High-Sensitivity DNA kit (Agilent Technologies, USA) prior to sequencing. DNA-libraries were sequenced using an Illumina HiSeq 1500 platform (Illumina, USA) with 150-bp paired-end reads.
Sequencing analysis
DNA-reads were mapped to the reference human genome (hg19) using the TopHat program. The differential gene expression between BA10L and BA10R were analyzed using the Cufflinks package from the Cufflinks program (Trapnell et al., 2012). RPKM analysis (reads per kilobase per million mapped reads) was used as the normalized value of the expression level (Mortazavi et al., 2008).
Gene Ontology (GO) analysis was performed using the DAVID 6.8 (Database for Annotation, Visualization and Integrated Discovery) (Huangda, Sherman, & Lempicki, 2009)and PANTHERclassification system (Mi et al., 2017). Cell type specific expression in the human cerebral cortex for studied genes was checked in the Human Protein Atlas database (version 4.1) and Ensembl (version 54.36) (Ponten et al., 2009).To investigate whether revealed genes with differential expression were constitutive or inducible, we used the database consisting of 3804 human housekeeping genes (Eisenberg, & Levanon, 2013). Finally, human disease enrichment and inducibility analysis of revealed genes was performed up to 30.03.2017 using the PubMed database and Schizophrenia Gene Resource (SZGR) (Jia, Han, Zhao, Lu, & Zhao, 2017).
Results
The total number of raw reads generated for both BA10L and BA10R areas was from 18 to 30 million sequences per sample (NCBI Bioproject: PRJNA388140). At least 90% of reads were mapped to the reference genome (except one sample). The vast majority of identified genes did not differ in expression between BA10L and BA10R areas. But some genes we found demonstrated significant differential expression.
The comparative study of the gene expression in the right and left parts of FPC allowed us to identify 61 genes that showed lateralization: 40 genes were up-regulated in BA10R whereas only 21 genes were up-regulated in BA10L Therefore, gene expression generally increased in BA10R relative to BA10L. All statistically significant cases of gene differential expression between BA10L and BA10R are listed in Table 1. Interestingly, the non-coding RNA (i.e. expression suppressors) and protein-coding genes were equally up-regulated in BA10L (10 and 11 genes, respectively), whereas the expression of protein-coding genes strongly dominated in BA10R (5 and 35 genes, respectively) (the chi-squared statistic is 9.1589, df = 3, p ≤ 0.01).
Table 1. Statistically significant differential gene expressions in the human FPC
|
Gene |
Brodmann areas |
log2(fold change) |
p-value |
q-value |
|
HSPA7 |
BA10L vs. BA10R |
3,8592 |
0,00005 |
0,01981 |
|
RGS1 |
BA10L vs. BA10R |
2,66997 |
0,0001 |
0,037021 |
|
IPO9 |
BA10L vs. BA10R |
-2,10138 |
0,00005 |
0,01981 |
|
MIR34A+MIR34AHG |
BA10L vs. BA10R |
-2,25978 |
0,00005 |
0,01981 |
|
GAS5+SNORD80+SNORA103 |
BA10L vs. BA10R |
-1,31268 |
0,00005 |
0,01981 |
|
BAG3 |
BA10L vs. BA10R |
1,63766 |
0,00005 |
0,01981 |
|
ADM |
BA10L vs. BA10R |
1,89858 |
0,00005 |
0,01981 |
|
CD44 |
BA10L vs. BA10R |
1,48236 |
0,00005 |
0,01981 |
|
NPAS4 |
BA10L vs. BA10R |
2,51096 |
0,00005 |
0,01981 |
|
IFITM3 |
BA10L vs. BA10R |
1,36025 |
0,00005 |
0,01981 |
|
H19+MIR675 |
BA10L vs. BA10R |
-6,43921 |
0,00005 |
0,01981 |
|
USP15 |
BA10L vs. BA10R |
-2,95303 |
0,00005 |
0,01981 |
|
MIR331+MIR3685 |
BA10L vs. BA10R |
4,65469 |
0,00005 |
0,01981 |
|
TMCC3 |
BA10L vs. BA10R |
1,98614 |
0,00005 |
0,01981 |
|
FOS |
BA10L vs. BA10R |
1,65584 |
0,00005 |
0,01981 |
|
SERPINA3 |
BA10L vs. BA10R |
5,68007 |
0,00005 |
0,01981 |
|
MEG8+MIR370 |
BA10L vs. BA10R |
-2,52033 |
0,00005 |
0,01981 |
|
HAUS4 |
BA10L vs. BA10R |
-2,57307 |
0,00005 |
0,01981 |
|
MIR9-3HG |
BA10L vs. BA10R |
-1,10873 |
0,00005 |
0,01981 |
|
NFATC2IP |
BA10L vs. BA10R |
1,38301 |
0,00005 |
0,01981 |
|
MT1X |
BA10L vs. BA10R |
1,27587 |
0,0001 |
0,037021 |
|
Non-protein coding gene or gene fragment, no protein prediction available |
BA10L vs. BA10R |
inf |
0,00005 |
0,01981 |
|
TAOK1 |
BA10L vs. BA10R |
-2,56172 |
0,00005 |
0,01981 |
|
NSRP1 |
BA10L vs. BA10R |
1,01791 |
0,00005 |
0,01981 |
|
ITGB4 |
BA10L vs. BA10R |
1,93095 |
0,00005 |
0,01981 |
|
SLC14A1 |
BA10L vs. BA10R |
1,34783 |
0,0001 |
0,037021 |
|
AMH+SF3A2 |
BA10L vs. BA10R |
1,9429 |
0,00005 |
0,01981 |
|
CREB3L3 |
BA10L vs. BA10R |
4,66498 |
0,00005 |
0,01981 |
|
CCDC61 |
BA10L vs. BA10R |
-1,58857 |
0,00005 |
0,01981 |
|
TBC1D17 |
BA10L vs. BA10R |
-2,49842 |
0,00005 |
0,01981 |
|
ATF5 |
BA10L vs. BA10R |
-2,04998 |
0,00005 |
0,01981 |
|
REXO1 |
BA10L vs. BA10R |
1,50042 |
0,00005 |
0,01981 |
|
MIR24-2+LOC284454 |
BA10L vs. BA10R |
-4,07299 |
0,00005 |
0,01981 |
|
Carcinoembryonic antigen related cell adhesion molecule 1 (Ceacam1), transcript variant 3, mRNA |
BA10L vs. BA10R |
inf |
0,00005 |
0,01981 |
|
Ensemble predicted protein coding |
BA10L vs. BA10R |
-inf |
0,00005 |
0,01981 |
|
HOXD1 |
BA10L vs. BA10R |
-6,65318 |
0,00005 |
0,01981 |
|
CCNYL1 |
BA10L vs. BA10R |
3,20327 |
0,00005 |
0,01981 |
|
SLC11A1 |
BA10L vs. BA10R |
2,46311 |
0,00005 |
0,01981 |
|
NOP56 |
BA10L vs. BA10R |
2,57613 |
0,00005 |
0,01981 |
|
LAMA5 |
BA10L vs. BA10R |
3,46762 |
0,00005 |
0,01981 |
|
HMOX1 |
BA10L vs. BA10R |
1,83245 |
0,00005 |
0,01981 |
|
PISD |
BA10L vs. BA10R |
2,18628 |
0,00005 |
0,01981 |
|
PLCL2 |
BA10L vs. BA10R |
3,07933 |
0,00005 |
0,01981 |
|
TOP2B |
BA10L vs. BA10R |
-2,91264 |
0,00005 |
0,01981 |
|
ZBTB20 |
BA10L vs. BA10R |
1,49539 |
0,0001 |
0,037021 |
|
RUBCN |
BA10L vs. BA10R |
2,35736 |
0,00005 |
0,01981 |
|
ARHGAP24 |
BA10L vs. BA10R |
3,26243 |
0,00005 |
0,01981 |
|
MIR7978 |
BA10L vs. BA10R |
-2,39482 |
0,00005 |
0,01981 |
|
Non-protein coding gene or gene fragment, no protein prediction available |
BA10L vs. BA10R |
1,85045 |
0,00005 |
0,01981 |
|
HSPA1A |
BA10L vs. BA10R |
3,41616 |
0,00005 |
0,01981 |
|
AQP1 |
BA10L vs. BA10R |
2,56426 |
0,00005 |
0,01981 |
|
HSPB1 |
BA10L vs. BA10R |
1,81864 |
0,00005 |
0,01981 |
|
PDK4 |
BA10L vs. BA10R |
1,49654 |
0,00005 |
0,01981 |
|
FAM83H |
BA10L vs. BA10R |
5,24147 |
0,00005 |
0,01981 |
|
DGAT1 |
BA10L vs. BA10R |
2,09862 |
0,00005 |
0,01981 |
|
GADD45G |
BA10L vs. BA10R |
1,94797 |
0,00005 |
0,01981 |
|
MIR6724-4 |
BA10L vs. BA10R |
3,2094 |
0,00005 |
0,01981 |
|
STS |
BA10L vs. BA10R |
-1,8016 |
0,00005 |
0,01981 |
|
TSIX |
BA10L vs. BA10R |
-1,96152 |
0,00005 |
0,01981 |
|
MIR221 |
BA10L vs. BA10R |
-4,19108 |
0,00005 |
0,01981 |
|
Non-protein coding gene or gene fragment, no protein prediction available |
BA10L vs. BA10R |
-6,11342 |
0,00005 |
0,01981 |
TheGO analysis of the gene expression data provides organized terms to describe characteristics of gene products in three categories: biological processes, molecular functions, and cellular components. This analysis also revealed a significant lateralization between left and right PFC. Sharing functions of up-regulated genes ─ binding, transporter and catalytic activities ─ were complemented by signal transducer activity in BA10L and by receptor activity in BA10R (Figure 1A). GO analysis of protein classes demonstrated strong increase in diversity of protein classes up-regulated in BA10R compared to BA10L, extending the latter with calcium binding proteins, cell adhesion molecules, receptors, chaperones, extracellular matrix proteins and other classes known to be involved in neuronal plasticity (Baucum, 2017; Dzyubenko, Gottschling, & Faissner, 2016; Gyurko, Soti, Stetak, & Csermely, 2014; Sheng, Leshchyns'ka, & Sytnyk 2013) (Figure 1B).
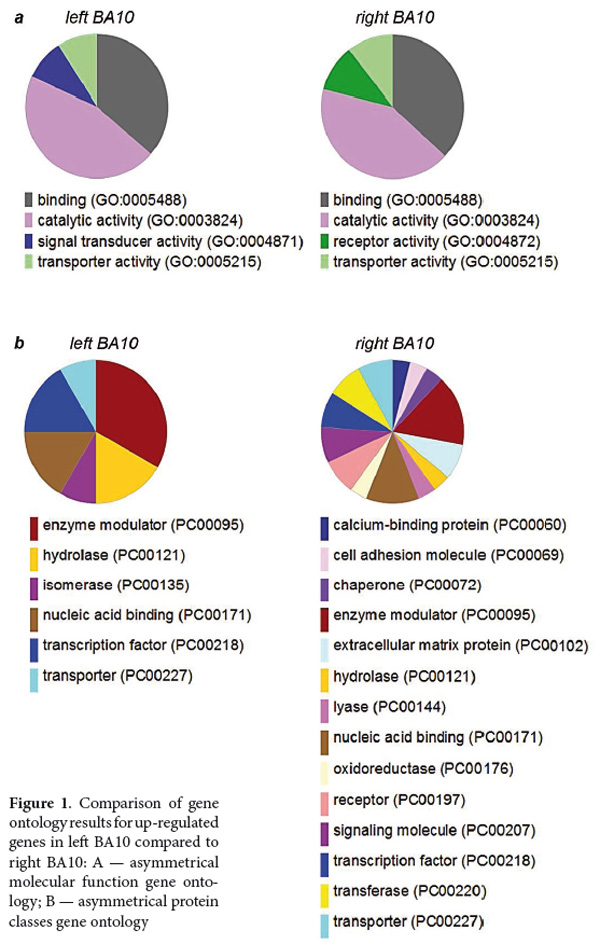
Further analysis of annotated protein expression in different cell types demonstrated the localization of revealed gene products in the neurons and neuropil. Among 61 studied genes, we found available information on annotated protein expression for 34 genes, with 31 genes showing neuronal expression and with 3 genes having expression in astrocytes and endothelial cells that regulate neuronal excitability and cerebral blood flow according to the activity of synapses (Bazargani, & Attwell, 2016; Nedoluzhko et al., in preparation).
To determine the functional significance of revealed differences between BA10L and BA10R, we also analyzed the data for the possible involvement of genes with differential expression in brain plasticity and activity-dependent mechanisms.In the group of neuronal genes with a strongly lateralized expression, it is worth noting C-FOSand NPAS4(Figure 2A, 2B,respectively). Both genes show rapid experience-dependent increase in expression levels and are widely used as markers of neuronal plasticity in functional molecular brain mapping in mice(Clayton, 2000; Ivashkina, Toropova, Ivanov, Chekhov, & Anokhin, 2016; X. Sun & Lin, 2016), and also in primate and human brain studies (Caston-Balderrama, Cameron, & Hoffman, 1998; Kaczmarek, Zangenehpour, & Chaudhuri, 1999; Nakagami, Watakabe, & Yamamori, 2013; Okuno & Miyashita, 1996; Rakhade et al., 2005). It is also necessary to note that two other genes, which are particularly strongly up-regulated in the right FPC, namely SERPINA3 and mir-331, are known for their role in molecular mechanisms of hippocampal formation. SERPINA3is an endothelial gene having a neurotrophic effect on hippocampal neurons (Kanai, Tanaka, & Hirai, 1991). Mir-33 is known to determine the expression of the neuropilin-2gene (Epis, Giles, Candy, Webster, & Leedman, 2014), a transmembrane receptor gene that regulates the dendritic spine density of pyramidal neurons (Demyanenko et al., 2014). Loss of neuropilin-2may induce aberrant processing within hippocampal and corticostriatal networks and thus contribute to neurodevelopmental disorders of memory and motor functions (Shiflett, Gavin, & Tran, 2015).
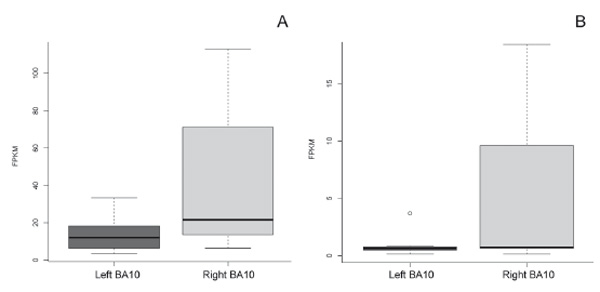
Figure 2. Examples of the relative increase in gene expression in left BA10 compared to right BA10: A – asymmetrical C-FOSexpression in BA10; B – asymmetricalNPAS4 expression in BA10
In the group of 21 genes with expression in the left FPC, we found only two genes with known functionality. One of them is the miR-34a, which inhibits expression of its prime target, sirtuin‑1(SIRT1), known to enhance cognitive abilities through proteostatic and neurotrophic mechanisms(Corpas et al., 2016, Lin, Mao, Song, & Huang, 2015).ATF5, a repressor of cyclic AMPinduced transcription (Pati, Meistrich, & Plon, 1999), is also up-regulated in BA10L compared with BA10R, whereas cAMPits molecular cascades have been shown to be central in regulating long-term memory and synaptic plasticity (Sheng, Leshchyns'ka, & Sytnyk, 2013).
By analyzing the published databases, we found information on induction-dependent gene expression for 47 of 61 genes in our dataset. Most of them (36 genes) showed inducible character of expression, whereas 11 genes expressed constitutively. There is a second approach to find inducible genes, which is to analyze the published database, consisting of 3804 human housekeeping genes, i.e. genes expected to be expressed irrespective of external signals (see Eisenberg, & Levanon, 2013). Among 21 genes up-regulated in BA10L, the database included four genes considered to be housekeeping (HAUS4, ARF5, TOP2B, IPO9). Among 40 genes with higher expression in BA10R, 5 genes were in the list of housekeeping genes (NSRP1, REXO1, NFATC2IP, NOP56, GADD45G). Thus, according to the second approach, up to 52 genes from the dataset of 61 differentially-expressed genes could be inducible.
Enrichment analysis of discovered genes with differential expression resulted in the detection of 30 genes implicated in mental disorders, and 15 genes related to other conditions, including non-mental brain disorders (gliomas, motor ataxia and others), with 16 genes omitted due to general absence of information on their role in the following categories: neurodevelopmental disorders (TOP2B, TAOK1, PISD), major depression (mir-34A, SPA1A, ARHGAP24), bipolar disorder (ATF5, SERPINA3), autism (STS, IFITM3), psychogenic stress (GAS5, SLC14A1), Alzheimer disease (PDK4, SLC11A1), Parkinson disease (RGS1), attention deficit hyperactivity disorder (STS) and frontotemporal dementia (NOP56). Strikingly, the vast majority of these differentially-expressed genes were implicated in pathogenesis of one single disorder: 24 out of 30 genes were disrupted in schizophrenia. These genes include mir-34A, mir-135B, ATF5, TOP2B, STS, SERPINA3, HSPA1A, RGS1, SLC11A1, IFITM3, MT1X, NSRP1, HSPB1, ZBTB20, C-FOS, BAG3, PDK4, PISD, ARHGAP24, PLCL2, ADM, GADD45G, CD44 and AQP1. The list of schizophrenia-related genes demonstrates a rightward asymmetry: 16 up-expressions in BA10R vs. only 8 in BA10L.
Discussion
In the present study, we applied the RNA-Seq technique to analyze lateralization of the molecular mechanisms in FPC, which have previously shown a diversity in anatomical and physiological features between left and right hemispheres, implicated in higher forms of cognitive processes, self-consciousness and voluntary control of behavior. For the first time, to our knowledge, a pronounced difference in molecular mechanisms of BA10L and BA10R was revealed. Until recently, it was often assumed thatgene expression in the cerebral cortex is bilaterally symmetrical (Hawrylycz et al., 2012; Pletikos et al., 2014). A new meta-analysis, which was conducted on microarray data, supposed that there are several examples of gene expression lateralization in the superior temporal cortex and auditory cortex of human adults (Karlebach, & Francks, 2015). However, these studies were based on only a small number of postmortem samples limited to brain areas with an obvious functional difference, namely language and speech processing.
Most genes with differential expression in BA10L and BA10R were found to be involved in brain plasticity and activity-dependent molecular mechanisms. Since we analyzed postnatal brain samples with a lack of information on functional cognitive load and with a broad range in postmortal interval, we assume that differential expression of inducible genes primarily reflects re-entry integrative mechanisms and remote cognitive processing (Edelman, & Gally, 2013)akin to activities within the Default Mode Network (DMN). For example, it is known that a baseline expression of immediate early genes is critical for off-line processing of cognitive information (Katche et al., 2010; Makino, Funayama, & Ikegaya, 2016). Moreover, the discovered dominance of inducible genes (about 80%) in our dataset of differential expression allows us to hypothesize that there is a hemispheric lateralization of the DMN in FPC. Recently, a similar lateralization of the effective connectivity has been discovered for a group of key DMN structures by extending dynamic causal modelling to hippocampal formation (Ushakov et al., 2016; Velichkovsky, Krotkova, Sharaev, & Ushakov, 2017).
Abouta half of the genes with lateralized expression in BA10 were related to mental disorders, wherein the vast majority (80%) were found to be disrupted in schizophrenia. Most of these schizophrenia-related genes (~67%) were up-expressed in BA10R. It can be assumed that FPC has a key involvement in this disease. No such wide association with other mental disorders was found. Schizophrenia is characterized by reduced hemispheric asymmetry of functional brain networks, as shown by recent connectome studies (Y. Sun etal., 2017). On the anatomical level, a similar conclusion was recently made with respect to autism spectrum disorder(Carper, Treiber, DeJesus, & Muller, 2016). Thus, lateralized expression of revealed genes seems to be critical for normal brain functioning.
To date, there is only one comparison of gene expression performed in brain tissue separately from the left and right hemispher areas (Mladinov et al., 2016). These authors studied dorsolateral prefrontal cortex (DLPFC, BA46) and the medial part of the orbitofrontal cortex (MOFC, BA11/12). The only difference they found in normal subjects in the right hemisphere compared with the left hemisphere was in BA11/12, which showed an increased expression of theKAT7gene and a decreased expression of gene NONO. KAT7is a lysine acetyltransferase 7, which may be involved in neuronal plasticity (Feng et al., 2016; Singh & Thakur, 2017).NONOis a protein phosphatase 1, regulatory subunit 114, required for tuning inhibitory synapses (Mircsof et al., 2015). In contrast, we revealed 61 genes differential expressed between right and left BA10. The difference in results between these studies could be explained by two reasons. Firstly, there is a long postmortem delay (PMD) time in the study by Mladinov et al. (2016). Samples in their study had ~19 h mean PMD, whereas samples we used had ~12 h mean PMD. As most genes with differential expression are localized to neurons, and neuronal RNA degrades faster than other brain RNAs, we suggest that the shorter PMD in our study could be the critical issue for sensitivity of RNA detection. A second reason could be simply that transcriptomic differences are more pronounced in BA10 than in BA11/12 or BA46 due to differential roles of the left and right PFC (Craik et al., 1999; Grady, Luk, Craik, & Bialystok, 2015; Stuss, & Benson, 1986). This functional lateralization may be reflected in the expression of molecular mechanisms reported in the present study as well as in the Yakovlevian Torque phenomenon known for a long time (LeMay, 1999; Toga, & Thompson, 2003).
On a more conceptual level, the ROBBIA model (ROtman-Baycrest Battery to Investigate Attention) seems to be relevant (Stuss, & Alexander, 2007; Ambrosini, & Vallesi, 2016). The model proposes a prefrontal specialization of two distinct executive functions. One is the “left-lateralized”task-setting function, defined as the transient cognitive control needed to form task-relevant rules and suppress task-irrelevant operations. The other specialization is the “right-lateralized” monitoring function, which provides the cognitive control needed to actively maintain abstract representations by monitoring their status in relation to each other and to intended plans of behaviour. If the right BA10 is in a state of sustained activation in contrast to phasic interventions of the task-setting processes in the left FPC, then we can expect enhanced rightwards asymmetry in activity-dependent molecular mechanisms since they are a target and a probable instrument for enduring maintenance of representations. On the other hand, a relative (48% vs. 12%) and absolute (10 vs. 5 out of 21 and 40) predominance of the non-coding RNA (i.e. expression suppressors) in BA10L would be appropriate to support sporadic acts of selecting and changing domain-specific goals. Indeed, to succeed in multiple choice and selective attention, one needs to suppress task-irrelevant operations. Of course, our hypothesis about contrasting lateralization of protein-coding and regulatory molecular mechanisms has to be tested in direct studies of the microRNA expression within BA10L and BA10R (Nedoluzhko et al., in preparation).
As mentioned above, there is a similarity between this dissociation in FPC and lateralization of processes within DMN. Hippocampal formation and possibly amygdalae are of primary interest here in view of growing evidence for asymmetries in their functional and effective connections (Kerestes, Chase, Phillips, Ladouceur, & Eickhoff, 2017; Ushakov et al., 2016). Although these asymmetries seem to be a feature of the specifically human large-scale brain architecture, their molecular precursors may well be considered in animal study data such as a new-found left-right dissociation of hippocampal memory processes in the mouse (El-Gaby, Shipton, & Paulsen, 2015).
Conclusion
Here, we present results of the first transcriptomic analysis of differential gene expression in the left and right human FPC. A coherent picture of differences is reported, revealing 40 genes that are up-expressed in BA10R and 21 genes up-expressed in BA10L. Differential expression is not confined to the number of genes but to their specialization as well. This has been shown by an additional GO analysis whereby protein-coding genes were predominantly expressed in BA10R and demonstrated a strong increase in diversity of protein classes compared to BA10L. In contrast, a relatively large proportion of up-expressed non-coding RNA has been discovered in BA10L. The results of this study are also potentially of clinical relevance since about half of the discovered genes with lateralized expression in FPC are implicated in mental disorders, first of all in schizophrenia.
Our analysis also opens up several lines of future work. First of all, the sensitivity of such studies could be improved by changing its current design to sequence different brain regions of the same donors. It would be of interest to compare current data from the right FPC with those of the left occipital lobe and in this way possibly contribute to a solution of the Yakovlevian Torque riddle. Even more important is to produce molecular portrayals of the large-scale networks such as DMN with a focus on molecular mechanisms of the amygdalae and both hippocampi which began to be investigated from the point of view of their functional and effective connections ((Kerestes et al., 2017; Ushakov et al. 2016). We hope that future research of gene regulation (e.g. epigenetic studies) at a higher resolution and with a better understanding of large-scale networks will benefit from these early insights into the molecular base of the most anterior parts of the human brain.
Acknowledgements
This work was in part supported by the Russian Science Foundation (grants 14-28-00234 and 17-78-30029 aimed at investigation of self-referential cognition and semantic representations in the human brain, respectively).
References
Ambrosini, E., & Vallesi, A. (2016). Asymmetry in prefrontal resting-state EEG spectral power underlies individual differences in phasic and sustained cognitive control. Neuroimage, 124, 843-857.doi: 10.1016/j.neuroimage.2015.09.035
Baucum, A.J., 2nd. (2017). Proteomic analysis of postsynaptic protein complexes underlying neuronal plasticity. ACS Chemical Neuroscience, 8,689-701.doi:10.1021/acschemneuro.7b00008
Bazargani, N., & Attwell, D. (2016). Astrocyte calcium signaling: The third wave. Nature Neuroscience, 19,182-189.doi: 10.1038/nn.4201
Braver, T.S., & Bongiolatti, S.R. (2002). The role of frontopolar cortex in subgoal processing during working memory. Neuroimage, 15,523-536.doi: 10.1006/nimg.2001.1019
Burgess, P.W., Dumontheil, I., & Gilbert, S.J. (2007). The gateway hypothesis of rostral prefrontal cortex (area 10) function. Trends in Cognitive Sciences, 11,290-298.doi: 10.1016/j.tics.2007.05.004
Carper, R.A., Treiber, J.M., DeJesus, S.Y., & Muller, R.A. (2016). Reduced hemispheric asymmetry of white matter microstructure in autism spectrum disorder.Journal of the American Academy of Child and Adolescent Psychiatry, 55,1073-1080.doi: 10.1016/j.jaac.2016.09.491
Caston-Balderrama, A.L., Cameron, J.L., & Hoffman, G.E. (1998). Immunocytochemical localization of Fos in perfused nonhuman primate brain tissue: Fixation and antisera selection. Journal of Histochemistry and Cytochemistry, 46,547-556. doi:10.1177/002215549804600416
Christoff, K., & Gabrieli, J.D.E. (2000). The frontopolar cortex and human cognition: Evidence for a rostrocaudal hierarchical organization within the human prefrontal cortex. Psychobiology, 28,168-186.
Clayton, D.F. (2000). The genomic action potential. Neurobiology of Learning Memory, 74,185-216.doi: 10.1006/nlme.2000.3967
Colantuoni, C., Lipska, B.K., Ye, T., Hyde, T.M., Tao, R., Leek, J.T., Colantuoni, E.A., … Kleinman J.E. (2011).Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature, 478,519-523.doi: 10.1038/nature10524
Corpas, R., Revilla, S., Ursulet, S., Castro-Freire, M., Kaliman, P., Petegnief, V., Gimenez-Llort, M., . . . Sanfeliu, C. (2016). SIRT1 overexpression in mouse hippocampus induces cognitive enhancement through proteostatic and neurotrophic mechanisms. Molecular Neurobiology. doi: 10.1007/s12035-016-0087-9
Craik, F.I.M., Moroz, T.M., Moscovitch, M., Stuss, D.T., Winocur, G., Tulving, E., & Kapur, S. (1999). In search of the self: A positron emission tomography study. Psychological Science, 10,26-34.doi: 10.1111/1467-9280.00102
Demyanenko, G.P, Mohan, V., Zhang, X., Brennaman, L.H., Dharbal, K.E., Tran, T.S., … Maness, P.F. (2014). Neural cell adhesion molecule NrCAM regulates Semaphorin 3F-induced dendritic spine remodeling. Journal of Neuroscience, 34,11274-11287.doi: 10.1523/JNEUROSCI.1774-14.2014
Dzyubenko, E., Gottschling, C., & Faissner, A. (2016). Neuron-glia interactions in neural plasticity: Contributions of neural extracellular matrix and perineuronal nets. Neural Plasticity, 2016,5214961. doi: 10.1155/2016/5214961
Edelman, G.M., & Gally, J.A. (2013). Reentry: A key mechanism for integration of brain function. Frontiers in Integrative Neuroscience, 7,63. doi: 10.3389/fnint.2013.00063
Eisenberg, E., & Levanon, E.Y. (2013). Human housekeeping genes, revisited. Trends in Genetics, 29,569-574.doi: 10.1016/j.tig.2013.05.010
El-GabyM., Shipton, O.A., & Paulsen, O. (2015). Synaptic plasticity and memory: New insights from hippocampal left-right asymmetries. Neuroscientist, 21(5), 490-502. doi: 10.1177/1073858414550658.
Epis, M.R., Giles, K.M., Candy, P.A., Webster, R.J., & Leedman, P.J. (2014). miR-331-3p regulates expression of neuropilin-2 in glioblastoma. Journal of Neuro-oncology, 116,67-75.doi: 10.1007/s11060-013-1271-7
Feng, Y., Vlassis, A., Roques, C., Lalonde, M.E., Gonzalez-Aguilera, C., Lambert, J.P., … Groth, A. (2016). BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. The EMBO Journal, 35,176-192.doi: 10.15252/embj.201591293
Forgács, B., Lukács, Á., & Pléh, C. (2014). Lateralized processing of novel metaphors: Disentangling figurativeness and novelty. Neuropsychologia, 56,101-109.doi: 10.1016/j.neuropsychologia.2014.01.003
Grady, C.L., Luk, G., Craik, F.I., & Bialystok, E. (2015). Brain network activity in monolingual and bilingual older adults. Neuropsychologia, 66,170-181.doi: 10.1016/j.neuropsychologia.2014.10.042
Gyurko, D.M., Soti, C., Stetak, A., & Csermely, P. (2014). System level mechanisms of adaptation, learning, memory formation and evolvability: The role of chaperone and other networks. Currunt Protein and Peptide Science, 15,171-188.doi: 10.2174/1389203715666140331110522
Halpern, M.E., Gunturkun, O., Hopkins, W.D., & Rogers, L.J. (2005). Lateralization of the vertebrate brain: Taking the side of model systems. Journal of Neuroscience, 25,10351-10357.doi: 10.1523/JNEUROSCI.3439-05.2005
Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, … Jones AR. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489,391-399.doi: 10.1038/nature11405
Herbert, M.R., Ziegler, D.A., Deutsch, C.K., O'Brien, L.M., Kennedy, D.N., Filipek, P.A., … Caviness, V.S.Jr., (2005). Brain asymmetries in autism and developmental language disorder: A nested whole-brain analysis. Brain, 128,213-226.doi: 10.1093/brain/awh330
Hrvoj-Mihic, B., Bienvenu, T., Stefanacci, L., Muotri, A.R., & Semendeferi, K. (2013). Evolution, development, and plasticity of the human brain: from molecules to bones. Frontiers in Human Neuroscience, 7,707. doi: 10.3389/fnhum.2013.00707
Huang da, W., Sherman, B.T., & Lempicki, R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4,44-57.
Ivashkina, O.I., Toropova, K.A., Ivanov, A.A., Chekhov, S.A., & Anokhin, K.V. (2016). Waves of c-Fos and Arc Ppoteins expression in neuronal populations of the hippocampus in response to a single episode of new experience. Bulletin of Experimental Biology and Medicine, 160,729-732.doi: 10.1007/s10517-016-3296-3
Jia, P., Han, G., Zhao, J., Lu, P., & Zhao, Z. (2017). SZGR 2.0: A one-stop shop of schizophrenia candidate genes. Nucleic Acids Research, 45,D915-D924. doi:10.1093/nar/gkw902
Kaczmarek, L., Zangenehpour, S., & Chaudhuri, A. (1999). Sensory regulation of immediate-early genes c-fos and zif268 in monkey visual cortex at birth and throughout the critical period. Cerebral Cortex, 9,179-187.doi: 10.1093/cercor/9.2.179
Kanai, H., Tanaka, M., & Hirai, S. (1991). Alpha-1-Antichymotrypsin has a trophic effect on hippocampal-neurons invitro. Neuroscience Letters, 125,163-165.doi: 10.1016/0304-3940(91)90017-N
Kang, H.J., Kawasawa, Y.I., Cheng, F., Zhu, Y., Xu, X., Li, M., … Sestan, N. (2011). Spatio-temporal transcriptome of the human brain. Nature, 478,483-489.doi: 10.1038/nature10523
Karlebach, G., & Francks, C. (2015). Lateralization of gene expression in human language cortex. Cortex, 67,30-36.doi: 10.1016/j.cortex.2015.03.003
Katche, C., Bekinschtein, P., Slipczuk, L., Goldin, A., Izquierdo, I.A., Cammarota, M., & Medina, J.H. (2010). Delayed wave of c-Fos expression in the dorsal hippocampus involved specifically in persistence of long-term memory storage. Proceedings of the National Academy of Sciences of the United States of America, 107,349-354.doi: 10.1073/pnas.0912931107
Kerestes, R., Chase, H.W., Phillips, M.L., Ladouceur, C.D., & Eickhoff, S.B. (2017). Multimodal evaluation of the amygdala's functional connectivity. Neuroimage, 148,219-229.doi: 10.1016/j.neuroimage.2016.12.023
LeMay, M. (1999). Functional and anatomical asymmetries of the human brain. European Journal of Neurology, 6,79-85.doi: 10.1046/j.1468-1331.1999.610079.x
Lin, Q., Mao, Y.R., Song, Y.L., & Huang, D.F. (2015). MicroRNA-34a induces apoptosis in PC12 cells by reducing B-cell lymphoma 2 and sirtuin-1 expression.Molecular Medicine Reports, 12,5709-5714.doi: 10.3892/mmr.2015.4185
Makino, K., Funayama, K., & Ikegaya, Y. (2016). Spatial clusters of constitutively active neurons in mouse visual cortex. Anatomical Science International, 91,188-195.doi: 10.1007/s12565-015-0284-z
Mi, H., Huang, X., Muruganujan, A., Tang, H., Mills, C., Kang, D., & Thomas, P.D. (2017). PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research, 45,D183-D189.doi: 10.1093/nar/gkw1138
Miller, E.K., & Cohen, J.D. (2001). An integrative theory of prefrontal cortex function. Annual Review of Neuroscience, 24,167-202. doi: 10.1146/annurev.neuro.24.1.167
Mircsof, D., Langouet, M., Rio, M., Moutton, S., Siquier-Pernet, K., Bole-Feysot, C., … Colleaux L. (2015). Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects. Nature Neuroscience, 18,1731-1736.doi: 10.1038/nn.4169
Mladinov, M., Sedmak, G., Fuller, H.R., Babic Leko, M., Mayer, D., Kirincich, J., … Simic, G. (2016). Gene expression profiling of the dorsolateral and medial orbitofrontal cortex in schizophrenia. Translational Neuroscience, 7,139-150.doi: 10.1515/tnsci-2016-0021
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5, 621-628.doi: 10.1038/nmeth.1226
Nakagami, Y., Watakabe, A., & Yamamori, T. (2013). Monocular inhibition reveals temporal and spatial changes in gene expression in the primary visual cortex of marmoset. Frontiers in Neural Circuits, 7,43. doi: 10.3389/fncir.2013.00043
Nedoluzhko, A.V., Efimova, O.I., Kildyushov, E.M., Dolina, I.A., Sokolov, A.S., Ushakov, V.L., Khaitovich, P.E. . . . Velichkovsky, B.M. (in preparation). Sequencing frontopolar cortex: New insights into the molecular base of human cerebral asymmetry.
Okuno, H., & Miyashita, Y. (1996). Expression of the transcription factor Zif268 in the temporal cortex of monkeys during visual paired associate learning. European Journal of Neuroscience, 8,2118-2128.doi: 10.1111/j.1460-9568.1996.tb00733.x
Pati, D., Meistrich, M.L., & Plon, S.E. (1999). Human Cdc34 and Rad6B ubiquitin-conjugating enzymes target repressors of cyclic AMP-induced transcription for proteolysis. Molecular and Cellular Biology, 19,5001-5013.doi: 10.1128/MCB.19.7.5001
Penfield, W., & Evans, J. (1935). The frontal lobe in man: A clinical study of maximum removals. Brain, 58,115-133.doi: 10.1093/brain/58.1.115
Pletikos, M., Sousa, A.M., Sedmak, G., Meyer, K.A., Zhu, Y., Cheng, F. . . . Sestan, N. (2014). Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron, 81,321-332.doi: 10.1016/j.neuron.2013.11.018
Ponten, F., Gry, M., Fagerberg, L., Lundberg, E., Asplund, A., Berglund, L., … Uhlen, M. (2009). A global view of protein expression in human cells, tissues, and organs. Molecular Systems Biology, 5, 337. doi: 10.1038/msb.2009.93
Pujol, J., Lopez-Sala, A., Deus, J., Cardoner, N., Sebastian-Galles, N., Conesa, G., & Capdevila, A. (2002). The lateral asymmetry of the human brain studied by volumetric magnetic resonance imaging. Neuroimage, 17, 670-679.doi: 10.1006/nimg.2002.1203
Rakhade, S.N., Yao, B., Ahmed, S., Asano, E., Beaumont, T.L., Shah, A.K., … Loeb, J.A. (2005). A common pattern of persistent gene activation in human neocortical epileptic foci. Annals Neurology, 58,736-747.doi: 10.1002/ana.20633
Ramnani, N., & Owen, A.M. (2004). Anterior prefrontal cortex: Insights into function from anatomy and neuroimaging. Nature Reviews Neuroscience, 5,184-194.doi: 10.1038/nrn1343
Renteria, M.E. (2012). Cerebral asymmetry: A quantitative, multifactorial, and plastic brain phenotype. Twin Research and Human Genetics, 15, 401-413.doi: 10.1017/thg.2012.13
Semendeferi, K., Armstrong, E., Schleicher, A., Zilles, K., & Van Hoesen, G.W. (2001). Prefrontal cortex in humans and apes: A comparative study of area 10. American Journal of Physical Anthropology, 114,224-241.doi: 10.1002/1096-8644(200103)114:3<224::AID-AJPA1022>3.0.CO;2-I
Sheng, L., Leshchynska, I., & Sytnyk, V. (2013). Cell adhesion and intracellular calcium signaling in neurons. Cell Communication and Signaling, 11,94. doi: 10.1186/1478-811X-11-94.
Shiflett, M.W., Gavin, M., & Tran, T.S. (2015). Altered hippocampal-dependent memory and motor function in neuropilin 2-deficient mice. Translational Psychiatry, 5,e521. doi: 10.1038/tp.2015.17.
Singh, P., & Thakur, M.K. (2017). Histone deacetylase 2 inhibition attenuates downregulation of hippocampal plasticity gene expression during aging. Molecular Neurobiology. doi: 10.1007/s12035-017-0490-x
Soch, J., Deserno, L., Assmann, A., Barman, A., Walter, H., Richardson-Klavehn, A., & Schott, B.H. (2016). Inhibition of information flow to the default mode network during self-reference versus reference to others. Cerebral Cortex.doi: 10.1093/cercor/bhw206
Stuss, D.T., & Alexander, M.P. (2007). Is there a dysexecutive syndrome? Philosophical Transactions of the Royal Society B, 362,901-915.doi: 10.1098/rstb.2007.2096
Stuss, D.T., & Benson, D.F. (1986). The frontal lobes.New York: Raven Press.
Sun, T., Collura, R.V., Ruvolo, M., Walsh, & C.A. (2006). Genomic and evolutionary analyses of asymmetrically expressed genes in human fetal left and right cerebral cortex. Cerebral Cortex,16 Suppl 1,i18-25. doi: 10.1093/cercor/bhk026
Sun, X., & Lin, Y. (2016). Npas4: Linking neuronal activity to memory. Trends in Neurosciences, 39,264-275.doi: 10.1016/j.tins.2016.02.003
Sun, Y., Chen, Y., Collinson, S.L., Bezerianos, A., & Sim, K. (2017). Reduced hemispheric asymmetry of brain anatomical networks is linked to schizophrenia: A connectome study. Cerebral Cortex, 27,602-615.
Toga, A.W., & Thompson, P.M. (2003). Mapping brain asymmetry. Nature Reviews Neuroscience, 4, 37-48.doi: 10.1038/nrn1009
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, . . . Pachter, L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562-578.doi: 10.1038/nprot.2012.016
Ushakov, V.L., Sharaev, M.G., Kartashov, S.I., Zayyalova, V.V., Verkhlyutov, V.M., & Velichkoysky, B.M. (2016). Dynamic Causal Modeling of hippocampal links within the Human Default Mode network: Lateralization and computational stability of effective connections. Frontiers in Human Neuroscience, 10,528. doi: 10.3389/fnhum.2016.00528
Velichkovsky, B. M., Krotkova, O. A., Sharaev, M. G., & Ushakov, V. L. (2017). In search of the “I”: Neuropsychology of lateralized thinking meets Dynamic Causal Modeling.Psychology in Russia: State of the Art, 10(3), 7-27. doi: 10.11621/pir.2017.0301
To cite this article: Dolina I.A., Efimova O.I..., Velichkovsky B.M. (2017). Exploring terra incognita of cognitive science: Lateralization of gene expression at the frontal pole of the human brain. Psychology in Russia: State of the Art, 10 (3), 231-247
The journal content is licensed with CC BY-NC “Attribution-NonCommercial” Creative Commons license.









